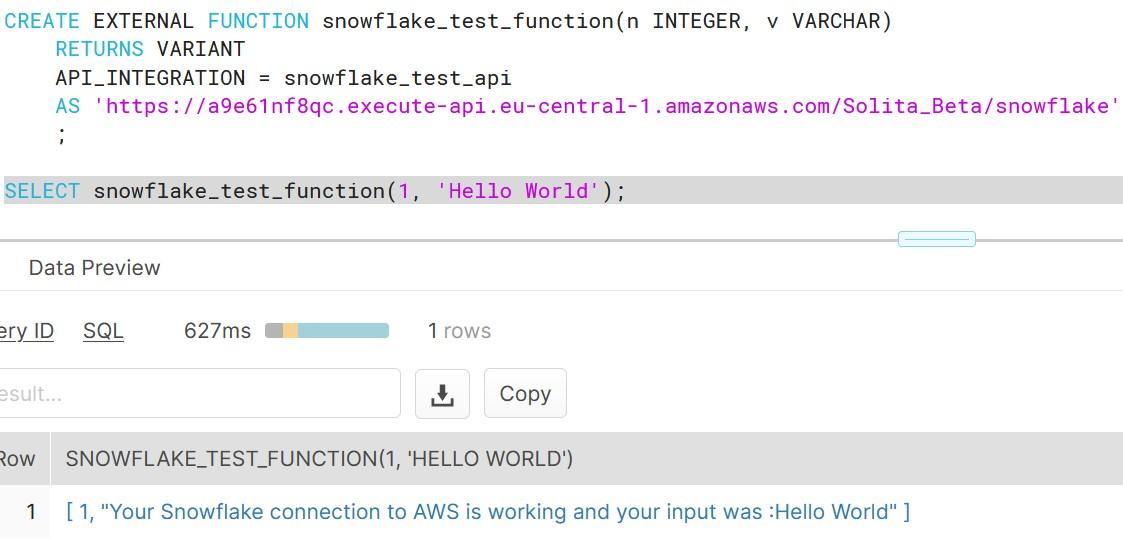
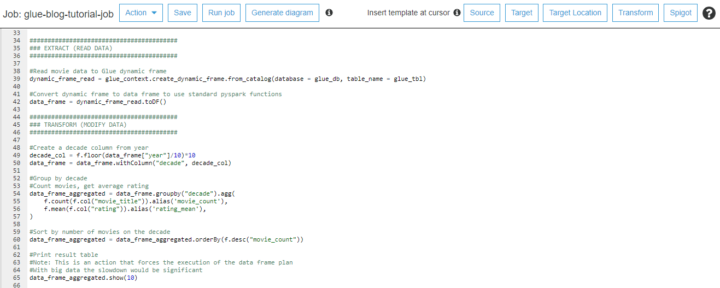
We have a miniseries before Christmas coming where we talk S-Q-L, /ˈsiːkwəl/ “sequel”. Yes, the 47 years old domain-specific language used in programming and designed for managing data. It’s very nice to see how old faithful SQL is going stronger than ever for stream processing as well the original relational database management purposes.
What is data then and how that should be used ? Take a look on article written in Finnish “Data ei ole öljyä, se on lantaa”
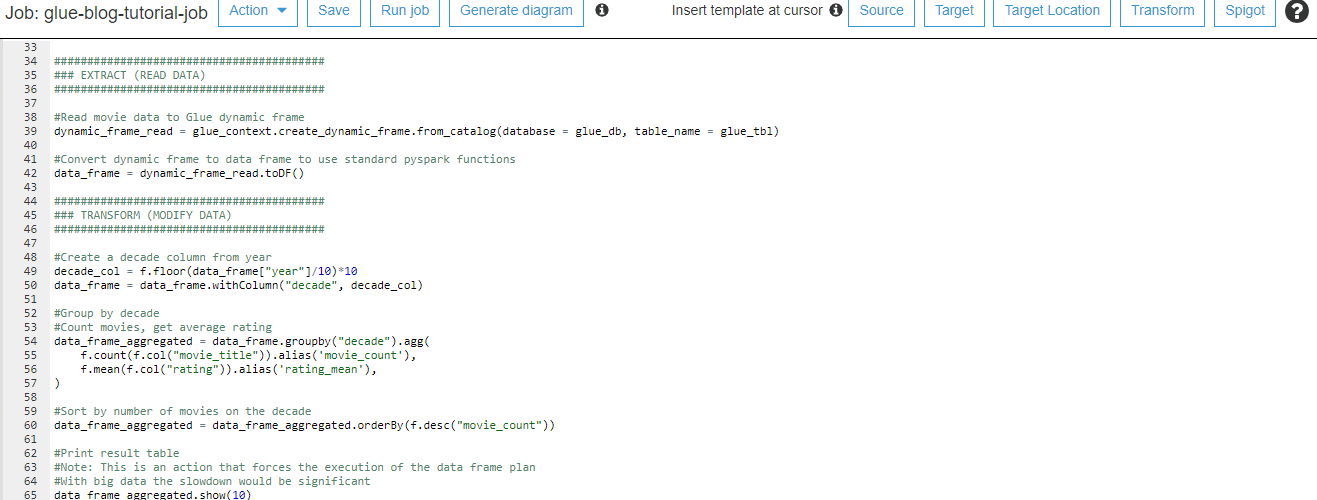
We will show you how to query and manipulate data across different solutions using the same SQL programming language.
The Solita Developer survey has become a tradition here at Solita and please check out the latest survey. It’s easy to see how SQL is dominating in a pool of many cool programming languages. It might take an average learner about two to three weeks to master the basic concepts of SQL and this is exactly what we will do with you.
Data modeling and real-time data
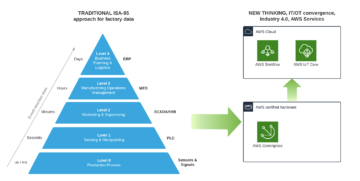
Operative technology (OT) solution have been real time from day one despite it’s also a question of illusion of real-time when it comes to IT systems. We could say that having network latency 5-15 ms towards Cloud and data processing with single-digit millisecond latency irrespective of the scale is considered near real time. This is important for Santa Claus and Industry 4.0 where autonomous fleet, robots and real-time processing in automation and control is a must have. Imagine situation where Santa’s autonomous sleigh with smart safety systems boosted computer vision (CV) able bypass airplanes and make smart decisions would have time of unit seconds or minutes – that would be a nightmare.
A data model is an abstract model that organizes elements of data and standardizes how they relate to one another and to the properties of real-world entities.
It’s easy to identify at least conceptual, logical and physical data models, where from the last one we are interested the most in this exercise to store and query data.
Back to the Future
Dimensional model heavily development by Ralph Kimball was breakthrough 1996 and had concepts like fact tables, dimension and ultimately creating a star schema. Challenge of this modeling is to keep conformed dimensions across the data warehouse and data processing can create unnecessary complexity.
One of the main driving factors behind using Data Vault is for both audit and historical tracking purposes. This methodology was developed by Daniel (Dan) Linstedt in early 2000. It has gain a lot of attraction being able to support especially modern cloud platform with massive parallel processing (MPP) of data loading and not to worry so much of which entity should be loaded first. Possibility even create data warehouse from scratch and just loading data in is pretty powerful when designing an idempotent system.
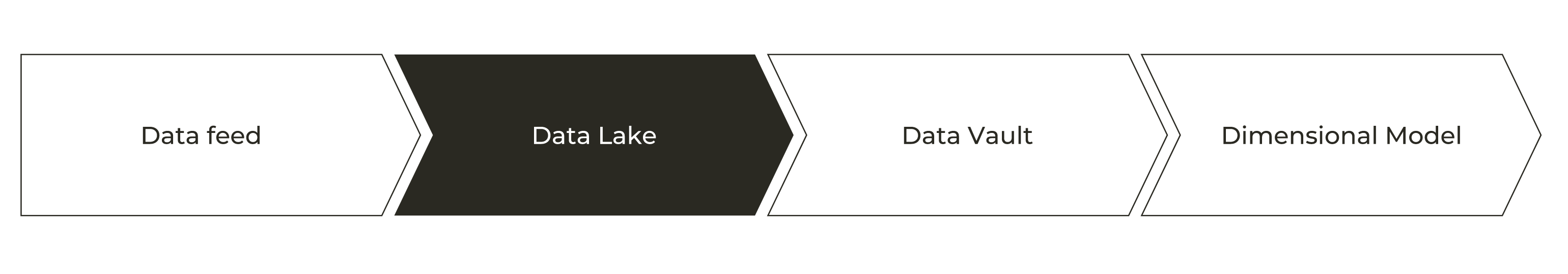
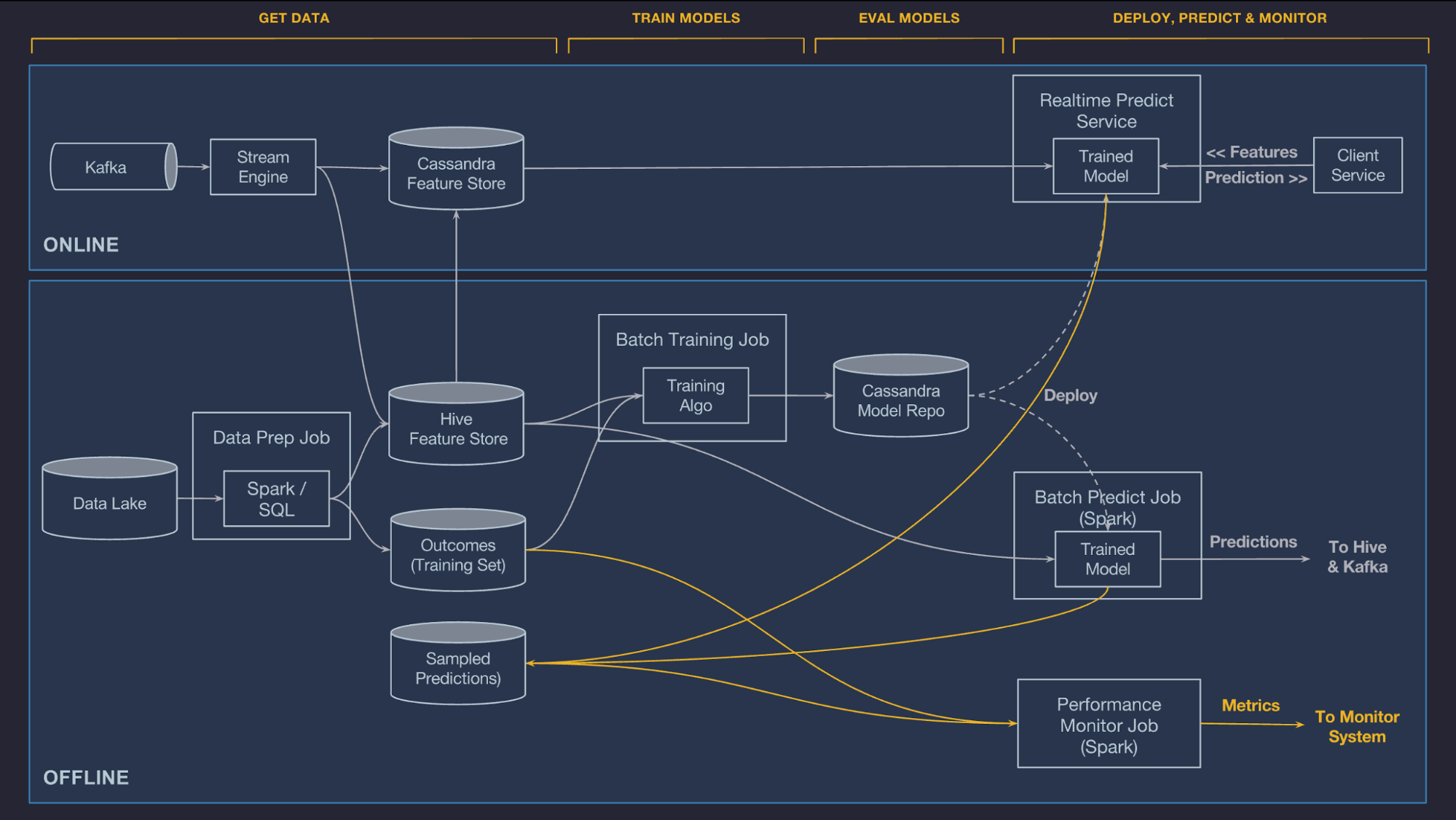
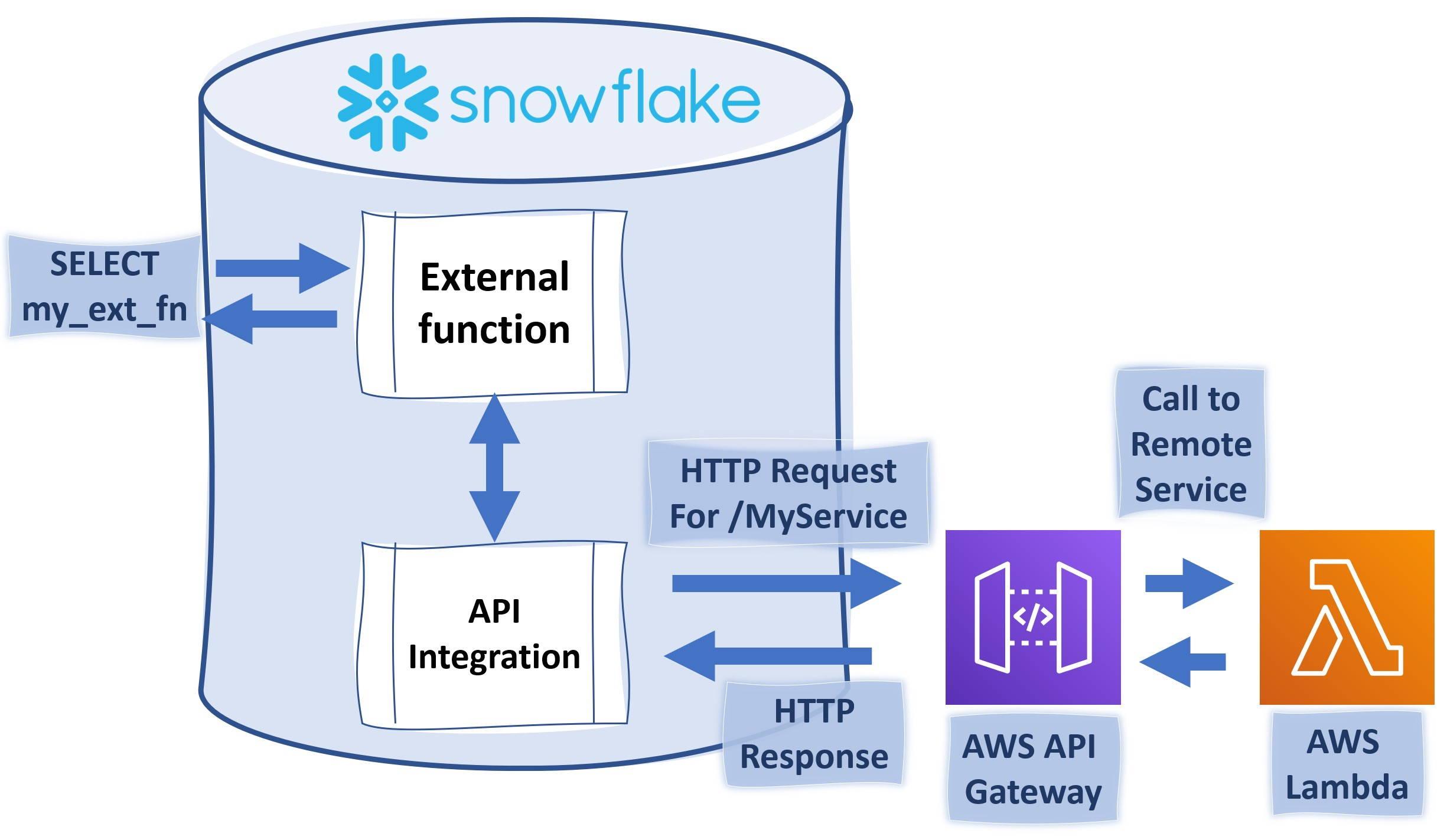
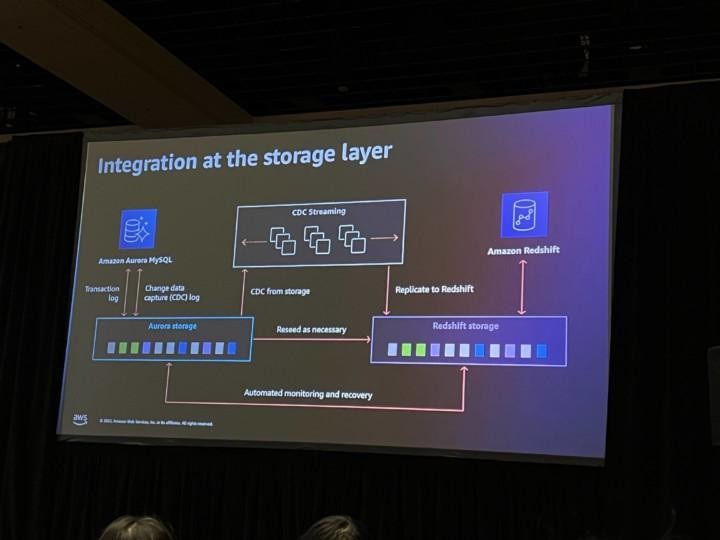
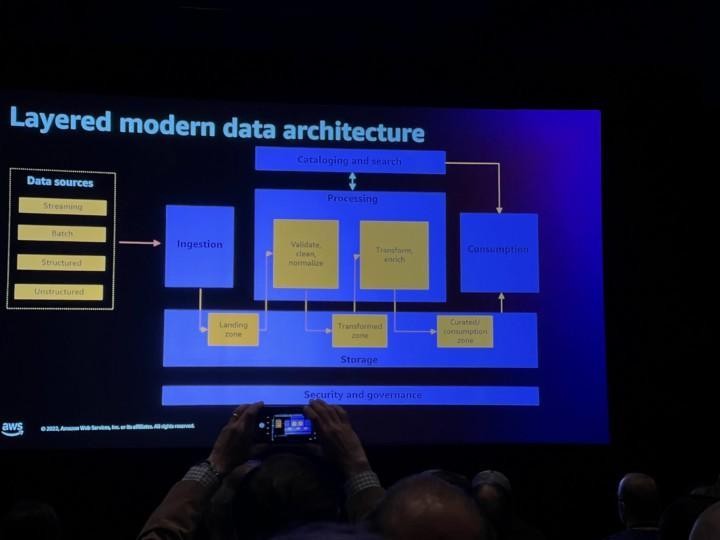
Quite typical data flow looks like picture above and like you already noticed this will have impact on how fast data is landed into applications and users. Theses for Successful Modern Data Warehousing are useful to read when you have time.
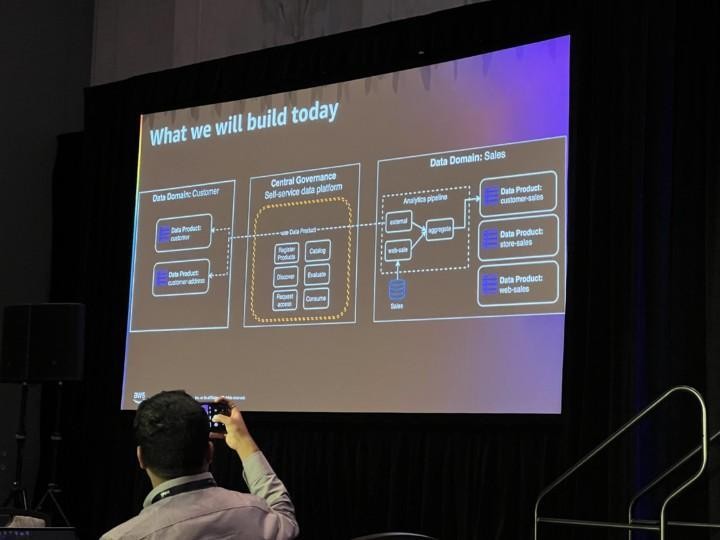
Data Mesh ultimate promise is to eliminate the friction to deliver quality data for producers and enable consumers to discover, understand and use the data at rapid speed. You could imagine this as data products in own sandboxes with some common control plane and governance. In any case to be successful you need expertise from different areas such as business, domain and data. End of the day Data Mesh does not take a strong position on data modeling.
Wide Tables / One Big Table (OBT) that is basically nested and denormalized tables is one modeling that is perhaps the mostly controversy. Shuffling data between compute instances when executing joins will have negative impact on performance (yes, you can e.g. replicate dimensional data to nodes and keep fact table distributed which will improve performance) and very often operational data structures produced by micro-services and exchanged over API are closer to this “nested” structure. Having same structure and logic for batch SQL as streaming SQL will ease your work.
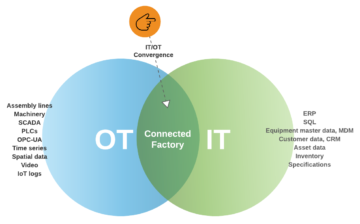
Breaking down the OT data items to multiple different sub optimal data strictures inside IT systems will loose the single, atomic data entity. Having said this it’s possible to ingest e.g. Avro files to MPP, keeping the structure same as original file as and using evolving schemas to discovery new attributes. That can be then use as baseline to load target layers such as Data Vault.
One interesting concept called Activity Schema that is sold us as being designed to make data modeling and analysis substantially simpler, faster.
Contextualize data
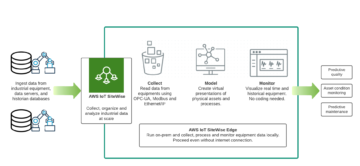
For our industrial Santa Claus case one very important thing is how to create inventory and contextualize data. One very promising path is an augmented data catalog that will cover a bit later. For some reason there is material out there explaining how IoT data has no structure which is just incorrect. The only reason I can think is that kind of data asset was not fit to traditional data warehouse thinking.
Something to take a look is Apache Avro that is a language-neutral data serialization system, developed by Doug Cutting, the father of Hadoop. The other one is JSON is an open standard file format and data interchange format that uses human-readable text to store and transmit data objects consisting of attribute–value pairs and arrays. This is not solution for data modeling even more you will notice later on this blog post how those are very valuable on steaming data and having schema compared to other formats like CSV.
Business case for Santa

Like always everything starts with Why and solution discovery phase, what we actual want to build and would that have a business value. At Christmas time our business is around gifts and how to deliver those on time. Our model is a bit more simplified and will include operational technology systems such as assets (Santa’s workshop) and fleet (sleighs) operations. There might always be something broken so few maintenance needs are pushed to technicians (elfs). Distributed data platform is used for supply chain and logistics analytics to remove bottlenecks so business owners can be satisfied (Santa Claus and the team) and all gifts will be delivered to the right address just in time.
Case Santa’s workshop
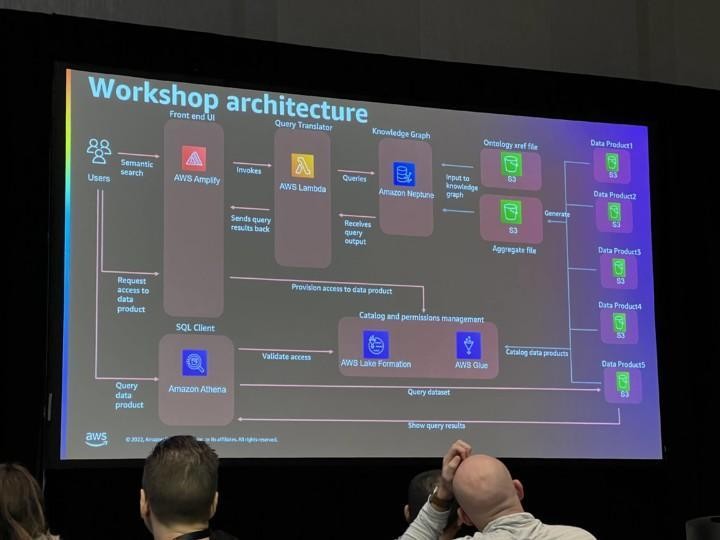
We can later use OEE to calculate that workshop performance in order to produce high quality nice gifts. Data is ingested real time and contextualized so once a while Santa and the team will check how we are doing. In this specific case we know that using Athena we can find relevant production line data just querying the S3 bucket where all raw data is stored already.
Day 1 – creating a Santa’s table for time series data
Let’s create a very basic table to capture all data from Santa’s factory floor. You will notice there are different data types like bigint and string. You can even add comments to help others to later find what kind of data field should include. In this case raw data is Avro but you do not have to worry about that so let’s go.
CREATE EXTERNAL TABLE `raw`(
`seriesid` string COMMENT 'from deserializer',
`timeinseconds` bigint COMMENT 'from deserializer',
`offsetinnanos` bigint COMMENT 'from deserializer',
`quality` string COMMENT 'from deserializer',
`doublevalue` double COMMENT 'from deserializer',
`stringvalue` string COMMENT 'from deserializer',
`integervalue` int COMMENT 'from deserializer',
`booleanvalue` boolean COMMENT 'from deserializer',
`jsonvalue` string COMMENT 'from deserializer',
`recordversion` bigint COMMENT 'from deserializer'
) PARTITIONED BY (
`startyear` string, `startmonth` string,
`startday` string, `seriesbucket` string
)
Day 2 – query Santas’s data
Now we have a table and how to query that one ? That is easy with SELECT and taking all fields using asterix. It’s even possible to limit that to 10 rows which is always a good practice.
SELECT * FROM "sitewise_out"."raw" limit 10;
Day 3 – Creating a view from query
View is a virtual presentation of data that will help to organize assets more efficiently. One golden rule is still now to create many views on top of other views and keep the solution simple. You will notice that CREATE VIEW works nicely and now we have timeinseconds and actual factory floor value (doublevalue) captured. You can even drop the view using DROP command.
CREATE OR REPLACE VIEW "v_santa_data"
AS SELECT timeinseconds, doublevalue FROM "sitewise_out"."raw" limit 10;
Day 4 – Using functions to format dates to Santa
You noticed that timeinseconds is in Epoch so let’s use functions to have more human readable output. So we add a small from_unixtime function and combine that with date_format to have formatted output like we want. Perfect, now we know from which data Santa Claus manufacturing data originated.
SELECT date_format(from_unixtime(timeinseconds),'%Y-%m-%dT%H:%i:%sZ') , doublevalue FROM "sitewise_out"."raw" limit 10;
Day 5 – CTAS creating a table
Using CTAS (CREATE TABLE AS SELECT) you can even create a new physical table easily. You will notice that Athena specific format has been added that you do not need on relational databases.
CREATE TABLE IF NOT EXISTS new_table_name
WITH (format='Avro') AS
SELECT timeinseconds , doublevalue FROM "sitewise_out"."raw" limit 10;
Day 6 – Limit the result sets
Now I want to limit the results to only those where the quality is Good.Adding a WHERE clause I can have only those rows printed to my output – that is cool!
SELECT * FROM "sitewise_out"."raw" where quality='GOOD' limit 10;
Case Santa’s fleet

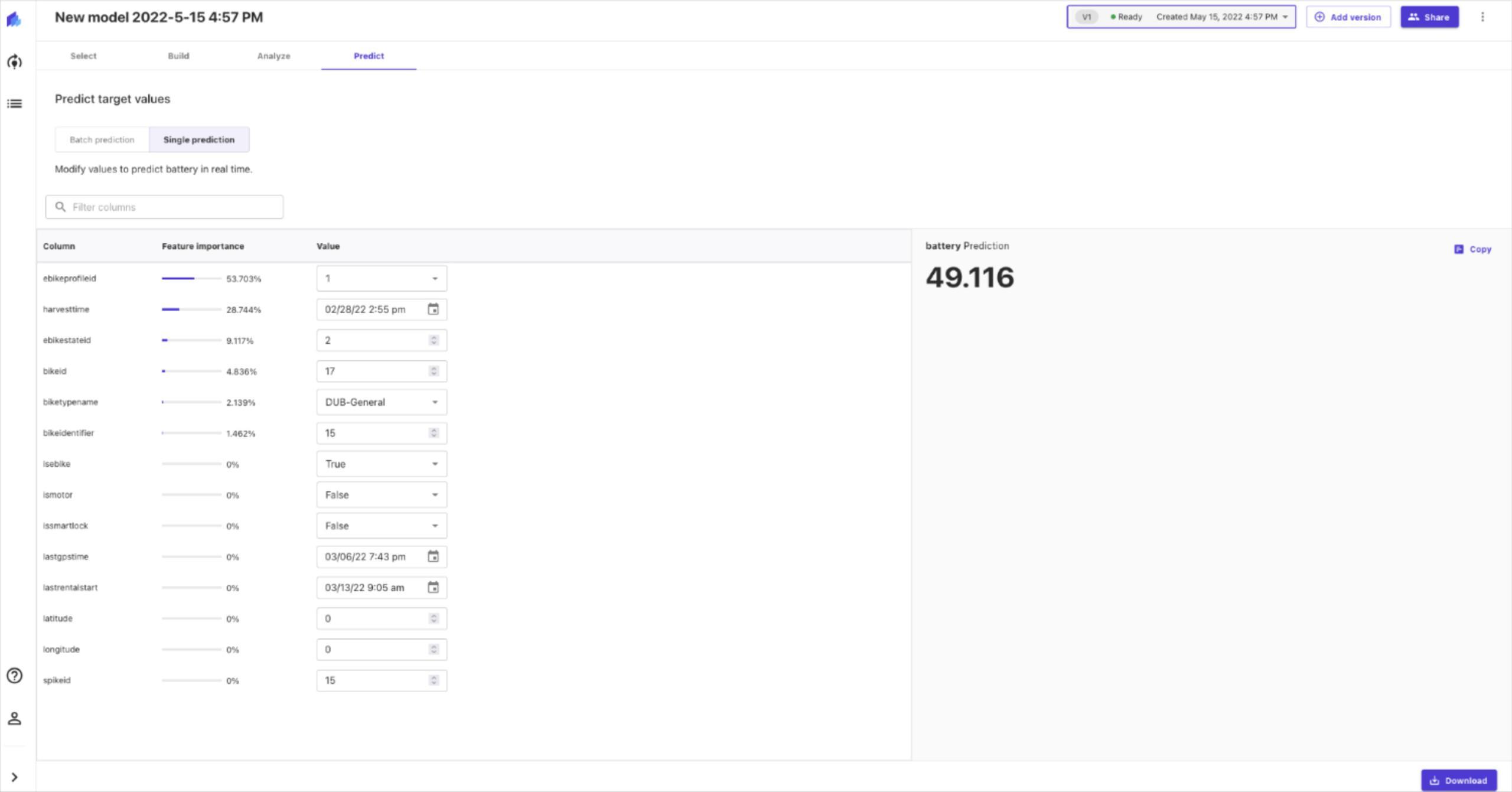
Now we jump into Santa’s fleet meaning sleights and there is few attribute we are interested like SleightD , IsSmartLock, LastGPSTime , SleightStateID , Latitude and Longitude. This data is time series that is ingested into our platform near real-time. Let’s use AWS Timestream service which is fast, scalable, and serverless time series database service for IoT and operational applications. A time series is a data set that tracks a sample over time.
Day 7 – creating a table for fleet
You will notice very quickly that data model looks different than on relational database cases. There is no need beforehand to define table structure just executing CreateTable is enough.
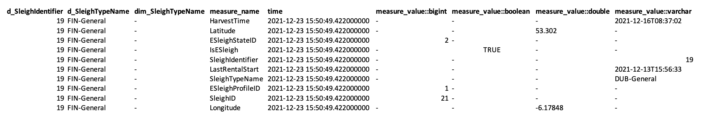
Day 8- query the latest record
You can override time field using e.g. LastGPSTime, in this example we use time when data was ingested in, so getting the last movement of sleigh would be like this.
SELECT * FROM movementdb.tbl_movement
ORDER BY time DESC
LIMIT 1
Day 9- let’s check the last 24 hours movement
We can use time to filter our results and ordering on descending same time.
SELECT *
FROM "movementdb"."tbl_movement"
WHERE time > ago(24h)
ORDER BY time DESC
Day 10- latitude and longitude
We can find out latitude and longitude information easily and please note we are using IN operator to bet both to query result.
SELECT measure_name,measure_value::double,time
FROM "movementdb"."tbl_movement"
WHERE time > ago(24h)
and measure_name in ('Longitude','Latitude')
ORDER BY time DESC LIMIT 10
Day 11- last connectivity info
Now we use 2 things so we group data based on sleigh id and find the maximum value. This will tell when sleigh was connected and sending data to our platform. There are plenty of functions to choose from so please check documentation.
SELECT greatest (time) as last_time, sleighId
FROM "movementdb"."tbl_movement"
WHERE time > ago(24h)
and measure_name = ('LastGPSTime')
group by sleighId,greatest (time)
Day 12- using conditions for smart lock data
CASE is very powerful to manipulate the query results so in this example we use that do indicate better if sleigh had smart lock.
SELECT time, measure_name,
CASE
WHEN measure_value::boolean = true THEN 'Yes we have a smart lock'
ELSE 'No we do not that kind of fancy locks'
END AS smart_lock_info
FROM "movementdb"."tbl_movement"
WHERE time between ago(1d) and now()
and measure_name='IsSmartLock'
Day 13- finding the latest battery level on each fleet equipment
This would be a bit more complex so we have one query to find max value of battery level and then we later join that to base data so on each record we know the latest battery level in the past 24 hours. Please notice we are using INNER join in this example.
WITH latest_battery_time as (
select
d_sleighIdentifier,
max(time) as latest_time
FROM
"movementdb"."tbl_movement"
WHERE
time between ago(1d)
and now()
and measure_name = 'Battery'
group by
d_sleighIdentifier
)
SELECT
b.d_sleighIdentifier,
b.measure_value :: double as last_battery_level
FROM
latest_battery_time a
inner join "movementdb"."tbl_movement" b on a.d_sleighIdentifier = b.d_sleighIdentifier
and b.time = a.latest_time
WHERE
b.time between ago(1d)
and now()
and b.measure_name = 'Battery'
Day 14- distinct values
The SELECT DISTINCT statement is used to return only distinct (different) values. This is so create and also very misused when removing duplicates etc. when actual problem can be on JOIN conditions.
SELECT
DISTINCT (d_sleighIdentifier)
FROM
"movementdb"."tbl_movement"
Day 15- partition by is almost magic
The PARTITION BY clause is a subclause of the OVER clause. The PARTITION BY clause divides a query’s result set into partitions. The window function is operated on each partition separately and recalculate for each partition. This is almost a magic and that can be used in several ways like in this example identify last sleigh Id.
select
d_sleighIdentifier,
SUM(1) as total,
from
(
SELECT
*,
first_value(d_sleighIdentifier) over (
partition by d_sleighTypeName
order by
time desc
) lastaction
FROM
"movementdb"."tbl_movement"
WHERE
time between ago(1d)
and now()
)
GROUP BY
d_sleighIdentifier,
lastaction
Day 16- interpolation (values of missing data points)
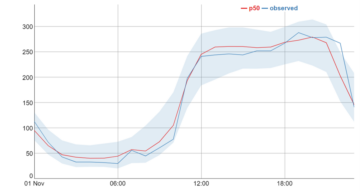
Timestream and few other IoT services supports linear interpolation, enabling to estimate and retrieve the values of missing data points in their time series data. This will come very handy when our fleet is not connected all the time, in this example we used it for our smart sleight battery level.
WITH rawseries as (
select
measure_value :: bigint as value,
time as d_time
from
"movementdb"."tbl_movement"
where
measure_name = 'Battery'
),
interpolate as (
SELECT
INTERPOLATE_LINEAR(
CREATE_TIME_SERIES(d_time, value),
SEQUENCE(
min(d_time),
max(d_time),
1s
)
) AS linear_ts
FROM
rawseries
)
SELECT
time,
value
FROM
interpolate CROSS
JOIN UNNEST(linear_ts)
Case Santa’s master data
 Now we jump into Master Data when factory and fleet is up are covered. In this very complex supply chain system customer data is very typical transactional data and in this exercise we keep it very atomic having stored only very basic info into DynamoDB that is a fully managed, serverless, key-value NoSQL database designed to run high-performance applications at any scale. We use this data to on IoT data streams for join, filtering and other purposes in fast manner. Good to remember that DynamoDB is not build for complex query patterns so it’s best on it’s original key=value data query pattern.
Now we jump into Master Data when factory and fleet is up are covered. In this very complex supply chain system customer data is very typical transactional data and in this exercise we keep it very atomic having stored only very basic info into DynamoDB that is a fully managed, serverless, key-value NoSQL database designed to run high-performance applications at any scale. We use this data to on IoT data streams for join, filtering and other purposes in fast manner. Good to remember that DynamoDB is not build for complex query patterns so it’s best on it’s original key=value data query pattern.
Day 17- adding master data
We upload our customer data into DynamoDB so called “items” based om the list received from Santa.
{
"customer_id": {
"S": "AJUUUUIIIOS"
},
"category_list": {
"L": [
{
"S": "Local Businesses"
},
{
"S": "Restaurants"
}
]
},
"homepage_url": {
"S": "it would be here"
},
"founded_year": {
"N": "2021"
},
"contract": {
"S": "NOPE"
},
"country_code": {
"S": "FI"
},
"name": {
"S": ""
},
"market_stringset": {
"SS": [
"Health",
"Wellness"
]
}
}
Day 18- query one customer item
Amazon DynamoDB supports PartiQL, a SQL-compatible query language, to select, insert, update, and delete data in Amazon DynamoDB. That is something we will use too speed up things. Let’s first query one customer data asset.
SELECT * FROM "tbl_customer" where customer_id='AJUUUUIIIOS'
Day 18- update kids information
Using the same PartiQL you can update item to have new attributes with one go.
UPDATE "tbl_customer"
SET kids='2 kids and one dog'
where customer_id='AJUUUUIIIOS'
Day 19- contains function
Now we can easily check that form marketing data who was interested on Health using CONTAINS. Many moderns database engines have native support for semi-structured data, including: Flexible-schema data types for loading semi-structured data without transformation. If you are not already familiar please take a look on AWS Redshift and Snowflake.
SELECT * FROM "tbl_customer" where contains("market_stringset", 'Health')
Day 20- inserting a new customer
Using familiar SQL like it’s very straightforward to add one new item.
INSERT INTO "tbl_customer" value {'name' : 'name here','customer_id' : 'A784738H'}
Day 21- missing data
Using a special MISSING you can find those where some attribute is not present easily.
SELECT * FROM "tbl_customer" WHERE "kids" is MISSING
Day 22- export data into s3
With one command you can export data from DynamoDB to S3 so let’s do that one based on documentation. AWS and others do have support for something called Federated Query where you can run SQL queries across data stored in relational, non-relational, object, and custom data sources. This federated feature we will cover later with You.
Day 23- using S3 select feature
Now you have data stored to S3 bucket and there is holder called /data so you can even use SQL to query S3 stored data. This will find relevant information for customer_id.
Select s.Item.customer_id from S3Object s
Day 24- s3 select to find right customer
You can even use customer Id to restrict data returned to you.
Select s.Item.customer_id from S3Object s where s.Item.customer_id.S ='AJUUUUIIIOS'
That’s all, I hope you get some glimpse how useful SQL is even you have different services and you might first think this will never be possible to use same kind of language of choice. Please do remember when some day You might be building next generation artificial intelligence and analysis platform with us knowing few data modeling techniques and SQL is a very good start.
You might be interested Industrial equipment data at scale for factory floor or manage your fleet at scale so let’s keep fresh mind and have a very nice week !
















































 Now we jump into Master Data when factory and fleet is up are covered. In this very complex supply chain system customer data is very typical transactional data and in this exercise we keep it very atomic having stored only very basic info into DynamoDB that
Now we jump into Master Data when factory and fleet is up are covered. In this very complex supply chain system customer data is very typical transactional data and in this exercise we keep it very atomic having stored only very basic info into DynamoDB that